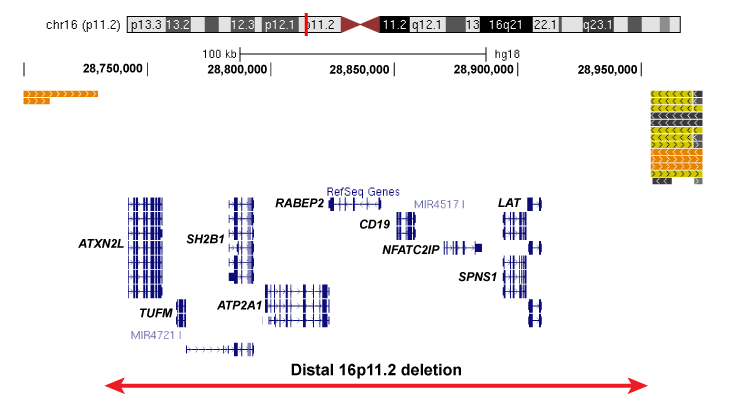
The distal 16p11.2 deletion is significantly associated with neurodevelopmental diseases and spans 9 genes of interest: ATXN2L, TUFM, SH2B1, ATP2A1, RABEP2, CD19, NFATC2IP, SPNS1, and LAT (right figure). The deletion is adjacent to the larger primary 16p11.2 deletion, which spans 25 genes including KCTD13, TAOK2, and MAPK3.
A wide array of clinical features have been associated with the distal 16p11.2 deletion. Developmental delay, intellectual disability and obesity are the most commonly-observed symptoms in individuals carrying the deletion. A subset of deletion carriers may also manifest autism spectrum disorders, behavioral problems such as ADHD, speech delay, schizophrenia, and skeletal defects such as scoliosis, while mild dysmorphic features and psychiatric disorders such as schizophrenia have also been observed in deletion carriers. Generally, individuals with the deletion do not share the same sets of symptoms, which indicates that this deletion is non-syndromic.
Individuals with the deletion have a sensitized genomic background rendering them vulnerable to multiple neurodevelopmental disorders such as intellectual disability, autism, and epilepsy. We are currently studying the distal 16p11.2 deletion as a paradigm to understand the complex genetics of developmental disorders. We hypothesize that genetic variants near or within the CNV interval and/or at a second site elsewhere in the genome contribute to the phenotypic outcome of disorders association with the deletion. Because the distal 16p11.2 deletion is inherited from parents in >70% of cases, assessing family history is essential to understanding the disease.
Most genomic deletions are the result of non-allelic homologous recombination (NAHR) between large (larger than 10kb) and highly similar sequences of DNA called segmental duplications. Several chromosomes are enriched in these types of DNA sequences, including the short arm of chromsome 16 (16p). This leads to a higher frequency of these rearrangements and therefore genomic disorders associated with this variation.
Study of the distal 16p11.2 deletion continues to be promising in understanding the variable clinical features associated with the disorder, and will lay a foundation to provide better treatment methods to patients in the years to come.
We are currently recruiting individuals with the distal 16p11.2 deletion and their family members for this study, and invite you to participate if your child or loved one has been affected by the deletion. The following link leads to an initial questionnaire, which we will use to obtain some preliminary information about you and your family members who carry the distal 16p11.2 deletion.
We estimate that the survey will take approximately 15 minutes to complete. We note that participation in this questionnaire is completely voluntary. All answers will be kept strictly confidential, and protected to the maximum extent as required by law.
After you complete the questionnaire, we will contact you directly with further information on our study. Your participation in this study will be very important in understanding the connections between genetic variants and the symptoms identified in individuals carrying the distal 16p11.2 deletion.
Link to 16p11.2 distal deletion questionnaire

